The dplyr package provides functions that perform data manipulation operations oriented to explore and manipulate datasets. At the most basic level, the package functions refers to data manipulation “verbs” such as select, filter, mutate, arrange, summarize among others that allow to chain multiple steps in a few lines of code. Currently dplyr supports four types of mutating joins, two types of filtering joins, and a nesting join. Mutating joins combine variables from the two data.frames: innerjoin return all rows from x where there are matching values in y, and all columns from x and y. Mtcars%% dplyr::filter(mpg30) making a new variable: mtcars% dplyr::mutate(efficient = ifelse(mpg30, TRUE, FALSE)) the pipe The variety of R syntaxes give you many ways to “say” the same thing read across the cheatsheet to see how different syntaxes approach the same problem.
Into R. Share plots, documents,. Spark MLlib and apps. H2O Extension Collect data into R for plotting Transformer function. dplyr verb. Direct Spark SQL (DBI). SDF function (Scala API). Export an R DataFrame. Read a file. Read existing Hive table Data Science in Spark with Sparklyr:: CHEAT SHEET Intro Using sparklyr. Manipulating Data with dplyr Overview. Dplyr is an R package for working with structured data both in and outside of R. Dplyr makes data manipulation for R users easy, consistent, and performant. With dplyr as an interface to manipulating Spark DataFrames, you can: Select, filter, and aggregate data.
data.table and dplyr cheat-sheet
This is a cheat-sheet on data manipulation using data.table and dplyr package (sqldf will be included soon…) . The package dplyr is an excellent and intuitive tool for data manipulation in R. Due to its intuitive data process steps and a somewhat similar concepts with SQL, dplyr gets increasingly popular. Another reason is that it can be integrated in SparkR seamlessly. Mastering dplyr will be a must if you want to get started with SparkR.
I found this cheat-sheet very useful in using dplyr. My post is inspired by it. I hereby write this cheat sheet for data manipulation with data.table / data.frame and dplyr computation side by side. It is especially useful for those who wants to convert data manipulation style from data.table to dplyr. There are 6 data investigation and manipulation included:
- Summary of data
- subset rows
- subset columns
- summarize data
- group data
- create new data
Select rows that meet logical criteria:
dplyr
data.frame / data.table
Remove duplicate rows:
dplyr
data.table
Randomly select fraction of rows
dplyr
Randomly select n rows
dplyr
data.table / data.frame
Select rows by position
dplyr
data.table / data.frame
Select and order top n entries (by group if group data)
dplyr Mac os catalina word 2011.
data.table
dplyr
data.frame
> iris[c(‘Sepal.Width’,’Petal.Length’,’Species’)]
data.table
Select columns whose name contains a character string
Select columns whose name ends with a character string
Select every column
dplyr
data.frame
Select columns whose name matches a regular expression
Select columns names x1,x2,x3,x4,x5
select(iris, num_range(‘x’, 1:5))
Select columns whose names are in a group of names
Select column whose name starts with a character string
Select all columns between Sepal.Length and Petal.Width (inclusive)
Select all columns except Species.
dplyr
data.frame
The package dplyr allows you to easily compute first, last, nth, n, n_distinct, min, max, mean, median, var, st of a vector as a summary of the table.
Summarize data into single row of values
dplyr
Apply summary function to each column
Note: mean cannot be applied on Factor type.
Count number of rows with each unique value of variable (with or without weights)
dplyr
data.table:
aggregate {stats}
Group data into rows with the same value of Species

dplyr
data.table: this is usually performed with some aggregation computation
Remove grouping information from data frame
dplyr
Compute separate summary row for each group
dplyr
data.frame
data.table
Mutate used window function, function that take a vector of values and return another vector of values, such as:
compute and append one or more new columns
data.frame / data.table
dplyr
Apply window function to each column
dplyr
base
data.table Latest version of mac os for macbook pro.
Compute one or more new columns. Drop original columns
Compute new variable by group.
dplyr
iris %>% group_by(Species) %>% mutate(ave = mean(Sepal.Length))
data.table
iris[, ave:=mean(Sepal.Length), by = Species]
data.frame
You can verify the result df1, df2 using:
Overview
QuestionsHow can I manipulate dataframes without repeating myself?
To be able to use the six main dataframe manipulation ‘verbs’ with pipes in
dplyr.To understand how
group_by()andsummarize()can be combined to summarize datasets.Be able to analyze a subset of data using logical filtering.
Manipulation of dataframes means many things to many researchers, we oftenselect certain observations (rows) or variables (columns), we often group thedata by a certain variable(s), or we even calculate summary statistics. We cando these operations using the normal base R operations:
But this isn’t very nice because there is a fair bit of repetition. Repeatingyourself will cost you time, both now and later, and potentially introduce somenasty bugs.
The dplyr package
Luckily, the dplyrpackage provides a number of very useful functions for manipulating dataframesin a way that will reduce the above repetition, reduce the probability of makingerrors, and probably even save you some typing. As an added bonus, you mighteven find the dplyr grammar easier to read.
Here we’re going to cover 6 of the most commonly used functions as well as usingpipes (%>%) to combine them.
select()filter()group_by()summarize()mutate()
If you have have not installed this package earlier, please do so:
Now let’s load the package:
Using select()
If, for example, we wanted to move forward with only a few of the variables inour dataframe we could use the select() function. This will keep only thevariables you select.
If we open up year_country_gdp we’ll see that it only contains the year,country and gdpPercap. Above we used ‘normal’ grammar, but the strengths ofdplyr lie in combining several functions using pipes. Since the pipes grammaris unlike anything we’ve seen in R before, let’s repeat what we’ve done aboveusing pipes.
To help you understand why we wrote that in that way, let’s walk through it stepby step. First we summon the gapminder dataframe and pass it on, using the pipesymbol %>%, to the next step, which is the select() function. In this casewe don’t specify which data object we use in the select() function since ingets that from the previous pipe. Fun Fact: There is a good chance you haveencountered pipes before in the shell. In R, a pipe symbol is %>% while in theshell it is | but the concept is the same!
Using filter()
If we now wanted to move forward with the above, but only with Europeancountries, we can combine select and filter
Challenge 1
Write a single command (which can span multiple lines and includes pipes) thatwill produce a dataframe that has the African values for lifeExp, countryand year, but not for other Continents. How many rows does your dataframehave and why?
Solution to Challenge 1
As with last time, first we pass the gapminder dataframe to the filter()function, then we pass the filtered version of the gapminder dataframe to theselect() function. Note: The order of operations is very important in thiscase. If we used ‘select’ first, filter would not be able to find the variablecontinent since we would have removed it in the previous step.
Using group_by() and summarize()
Now, we were supposed to be reducing the error prone repetitiveness of what canbe done with base R, but up to now we haven’t done that since we would have torepeat the above for each continent. Instead of filter(), which will only passobservations that meet your criteria (in the above: continent'Europe'), wecan use group_by(), which will essentially use every unique criteria that youcould have used in filter.
You will notice that the structure of the dataframe where we used group_by()(grouped_df) is not the same as the original gapminder (data.frame). Agrouped_df can be thought of as a list where each item in the listis adata.frame which contains only the rows that correspond to the a particularvalue continent (at least in the example above).
Using summarize()
The above was a bit on the uneventful side but group_by() is much moreexciting in conjunction with summarize(). This will allow us to create newvariable(s) by using functions that repeat for each of the continent-specificdata frames. That is to say, using the group_by() function, we split ouroriginal dataframe into multiple pieces, then we can run functions(e.g. mean() or sd()) within summarize().
That allowed us to calculate the mean gdpPercap for each continent, but it getseven better.
Challenge 2
Calculate the average life expectancy per country. Which has the longest average lifeexpectancy and which has the shortest average life expectancy?
Solution to Challenge 2
Another way to do this is to use the dplyr function arrange(), whicharranges the rows in a data frame according to the order of one or morevariables from the data frame. It has similar syntax to other functions fromthe dplyr package. You can use desc() inside arrange() to sort indescending order.
The function group_by() allows us to group by multiple variables. Let’s group by year and continent.
That is already quite powerful, but it gets even better! You’re not limited to defining 1 new variable in summarize().
count() and n()
A very common operation is to count the number of observations for eachgroup. The dplyr package comes with two related functions that help with this.
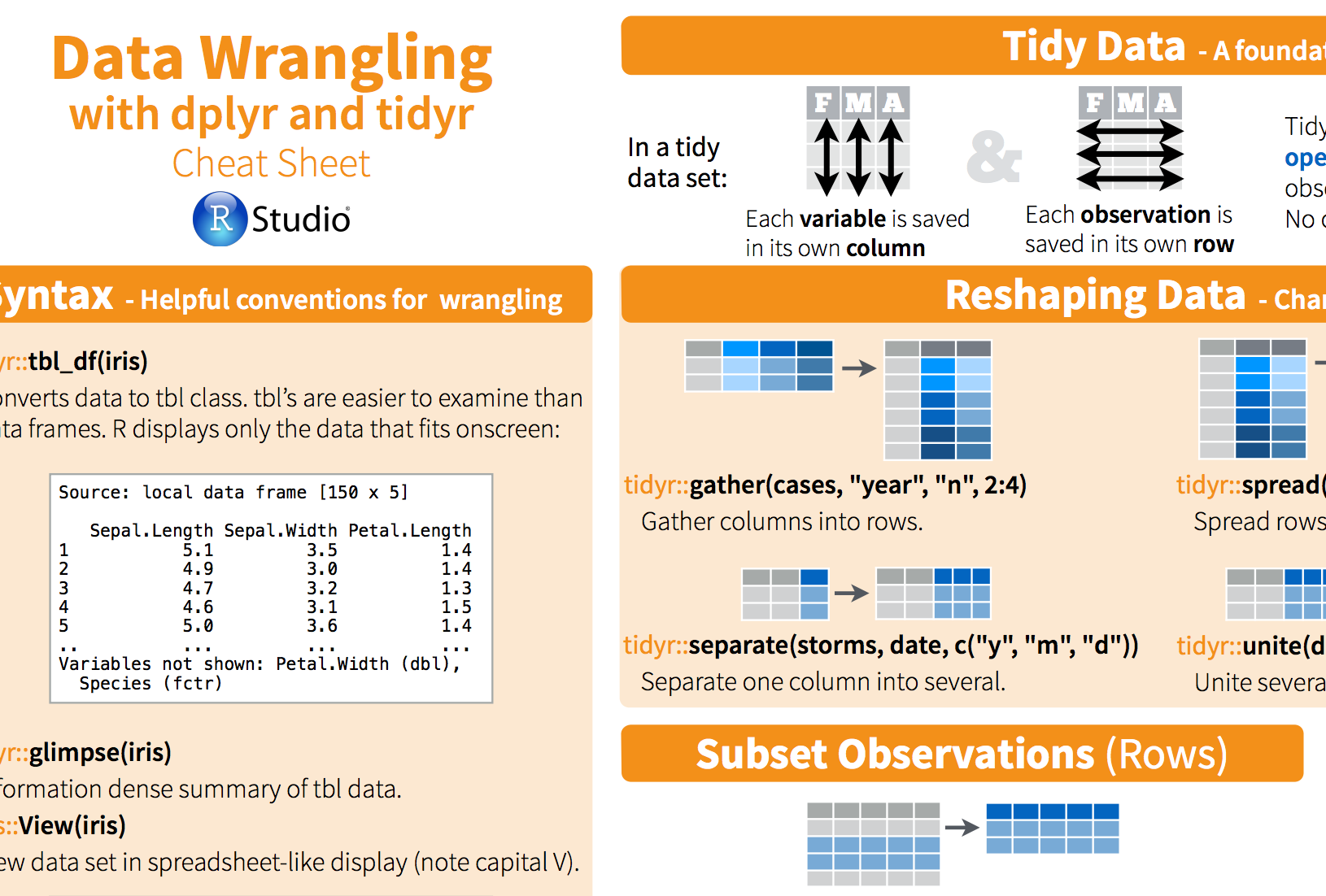
Tidyr Cheat Sheet
For instance, if we wanted to check the number of countries included in thedataset for the year 2002, we can use the count() function. It takes the nameof one or more columns that contain the groups we are interested in, and we canoptionally sort the results in descending order by adding sort=TRUE:
If we need to use the number of observations in calculations, the n() functionis useful. For instance, if we wanted to get the standard error of the lifeexpectency per continent:
You can also chain together several summary operations; in this case calculating the minimum, maximum, mean and se of each continent’s per-country life-expectancy:
Using mutate()
We can also create new variables prior to (or even after) summarizing information using mutate().
Connect mutate with logical filtering: ifelse
When creating new variables, we can hook this with a logical condition. A simple combination ofmutate() and ifelse() facilitates filtering right where it is needed: in the moment of creating something new.This easy-to-read statement is a fast and powerful way of discarding certain data (even though the overall dimensionof the data frame will not change) or for updating values depending on this given condition.
Combining dplyr and ggplot2
In the plotting lesson we looked at how to make a multi-panel figure by addinga layer of facet panels using ggplot2. Here is the code we used (with someextra comments):
This code makes the right plot but it also creates some variables (starts.withand az.countries) that we might not have any other uses for. Just as we used%>% to pipe data along a chain of dplyr functions we can use it to pass datato ggplot(). Because %>% replaces the first argument in a function we don’tneed to specify the data = argument in the ggplot() function. By combiningdplyr and ggplot2 functions we can make the same figure without creating anynew variables or modifying the data.
Using dplyr functions also helps us simplify things, for example we couldcombine the first two steps:
Advanced Challenge
Calculate the average life expectancy in 2002 of 2 randomly selected countriesfor each continent. Then arrange the continent names in reverse order.Hint: Use the dplyr functions arrange() and sample_n(), they havesimilar syntax to other dplyr functions.
Tibble Cheat Sheet
Solution to Advanced Challenge
R Studio Dplyr Cheat Sheet
Other great resources
Key Points
Dplyr In R Cheat Sheet
Use the
dplyrpackage to manipulate dataframes.Use
select()to choose variables from a dataframe.Cisco anyconnect ip address. Use
filter()to choose data based on values.Use
group_by()andsummarize()to work with subsets of data.Use
mutate()to create new variables.
